
Chapter 9: Advanced Data Wrangling With Pandas¶
Chapter Outline
Chapter Learning Objectives¶
Manipulate strings in Pandas by accessing methods from the
Series.strattribute.Understand how to use regular expressions in Pandas for wrangling strings.
Differentiate between datetime object in Pandas such as
Timestamp,Timedelta,Period,DateOffset.Create these datetime objects with functions like
pd.Timestamp(),pd.Period(),pd.date_range(),pd.period_range().Index a datetime index with partial string indexing.
Perform basic datetime operations like splitting a datetime into constituent parts (e.g.,
year,weekday,second, etc), apply offsets, change timezones, and resample with.resample().Make basic plots in Pandas by accessing the
.plotattribute or importing functions frompandas.plotting.
1. Working With Strings¶
import pandas as pd
import numpy as np
pd.set_option("display.max_rows", 20)
Working with text data is common in data science. Luckily, Pandas Series and Index objects are equipped with a set of string processing methods which we’ll explore here.
String dtype¶
String data is represented in pandas using the object dtype, which is a generic dtype for representing mixed data or data of unknown size. It would be better to have a dedicated dtype and Pandas has just introduced this: the StringDtype. object remains the default dtype for strings however, as Pandas looks to continue testing and improving the string dtype. You can read more about the StringDtype in the Pandas documentation here.
String Methods¶
We’ve seen how libraries like NumPy and Pandas can vectorise operations for increased speed and useability:
x = np.array([1, 2, 3, 4, 5])
x * 2
array([ 2, 4, 6, 8, 10])
This is not the case for arrays of strings however:
x = np.array(['Tom', 'Mike', 'Tiffany', 'Joel', 'Varada'])
x.upper()
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-3-acaefb05bf10> in <module>
1 x = np.array(['Tom', 'Mike', 'Tiffany', 'Joel', 'Varada'])
----> 2 x.upper()
AttributeError: 'numpy.ndarray' object has no attribute 'upper'
Instead, you would have to operate on each string object one at a time, using a loop for example:
[name.upper() for name in x]
['TOM', 'MIKE', 'TIFFANY', 'JOEL', 'VARADA']
But even this will fail if your array contains a missing value:
x = np.array(['Tom', 'Mike', None, 'Tiffany', 'Joel', 'Varada'])
[name.upper() for name in x]
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-5-b687bbdcc894> in <module>
1 x = np.array(['Tom', 'Mike', None, 'Tiffany', 'Joel', 'Varada'])
----> 2 [name.upper() for name in x]
<ipython-input-5-b687bbdcc894> in <listcomp>(.0)
1 x = np.array(['Tom', 'Mike', None, 'Tiffany', 'Joel', 'Varada'])
----> 2 [name.upper() for name in x]
AttributeError: 'NoneType' object has no attribute 'upper'
Pandas addresses both of these issues (vectorization and missing values) with its string methods. String methods can be accessed by the .str attribute of Pandas Series and Index objects. Pretty much all built-in string operations (.upper(), .lower(), .split(), etc) and more are available.
s = pd.Series(x)
s
0 Tom
1 Mike
2 None
3 Tiffany
4 Joel
5 Varada
dtype: object
s.str.upper()
0 TOM
1 MIKE
2 None
3 TIFFANY
4 JOEL
5 VARADA
dtype: object
s.str.split("ff", expand=True)
| 0 | 1 | |
|---|---|---|
| 0 | Tom | None |
| 1 | Mike | None |
| 2 | None | None |
| 3 | Ti | any |
| 4 | Joel | None |
| 5 | Varada | None |
s.str.len()
0 3.0
1 4.0
2 NaN
3 7.0
4 4.0
5 6.0
dtype: float64
We can also operate on Index objects (i.e., index or column labels):
df = pd.DataFrame(np.random.rand(5, 3),
columns = ['Measured Feature', 'recorded feature', 'PredictedFeature'],
index = [f"ROW{_}" for _ in range(5)])
df
| Measured Feature | recorded feature | PredictedFeature | |
|---|---|---|---|
| ROW0 | 0.963340 | 0.662353 | 0.862000 |
| ROW1 | 0.314565 | 0.169066 | 0.459403 |
| ROW2 | 0.929248 | 0.583589 | 0.689794 |
| ROW3 | 0.807835 | 0.940307 | 0.843171 |
| ROW4 | 0.865981 | 0.751341 | 0.812160 |
type(df.columns)
pandas.core.indexes.base.Index
Let’s clean up those labels by:
Removing the word “feature” and “Feature”
Lowercase the “ROW” and add an underscore between the digit and letters
df.columns = df.columns.str.capitalize().str.replace("feature", "").str.strip()
df.index = df.index.str.lower().str.replace("w", "w_")
df
| Measured | Recorded | Predicted | |
|---|---|---|---|
| row_0 | 0.963340 | 0.662353 | 0.862000 |
| row_1 | 0.314565 | 0.169066 | 0.459403 |
| row_2 | 0.929248 | 0.583589 | 0.689794 |
| row_3 | 0.807835 | 0.940307 | 0.843171 |
| row_4 | 0.865981 | 0.751341 | 0.812160 |
Great that worked! There are so many string operations you can use in Pandas. Here’s a full list of all the string methods available in Pandas that I pulled from the documentation:
Method |
Description |
|---|---|
|
Concatenate strings |
|
Split strings on delimiter |
|
Split strings on delimiter working from the end of the string |
|
Index into each element (retrieve i-th element) |
|
Join strings in each element of the Series with passed separator |
|
Split strings on the delimiter returning DataFrame of dummy variables |
|
Return boolean array if each string contains pattern/regex |
|
Replace occurrences of pattern/regex/string with some other string or the return value of a callable given the occurrence |
|
Duplicate values ( |
|
“Add whitespace to left, right, or both sides of strings” |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Split long strings into lines with length less than a given width |
|
Slice each string in the Series |
|
Replace slice in each string with passed value |
|
Count occurrences of pattern |
|
Equivalent to |
|
Equivalent to |
|
Compute list of all occurrences of pattern/regex for each string |
|
“Call |
|
“Call |
|
“Call |
|
Compute string lengths |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Return Unicode normal form. Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
|
Equivalent to |
I will also mention that I often use the dataframe method df.replace() to do string replacements:
df = pd.DataFrame({'col1': ['replace me', 'b', 'c'],
'col2': [1, 99999, 3]})
df
| col1 | col2 | |
|---|---|---|
| 0 | replace me | 1 |
| 1 | b | 99999 |
| 2 | c | 3 |
df.replace({'replace me': 'a',
99999: 2})
| col1 | col2 | |
|---|---|---|
| 0 | a | 1 |
| 1 | b | 2 |
| 2 | c | 3 |
Regular Expressions¶
A regular expression (regex) is a sequence of characters that defines a search pattern. For more complex string operations, you’ll definitely want to use regex. Here’s a great cheatsheet of regular expression syntax. I am self-admittedly not a regex expert, I usually jump over to RegExr.com and play around until I find the expression I want. Many Pandas string functions accept regular expressions as input, these are the ones I use most often:
Method |
Description |
|---|---|
|
Call re.match() on each element, returning a boolean. |
|
Call re.match() on each element, returning matched groups as strings. |
|
Call re.findall() on each element |
|
Replace occurrences of pattern with some other string |
|
Call re.search() on each element, returning a boolean |
|
Count occurrences of pattern |
|
Equivalent to str.split(), but accepts regexps |
|
Equivalent to str.rsplit(), but accepts regexps |
For example, we can easily find all names in our Series that start and end with a consonant:
s = pd.Series(['Tom', 'Mike', None, 'Tiffany', 'Joel', 'Varada'])
s
0 Tom
1 Mike
2 None
3 Tiffany
4 Joel
5 Varada
dtype: object
s.str.findall(r'^[^AEIOU].*[^aeiou]$')
0 [Tom]
1 []
2 None
3 [Tiffany]
4 [Joel]
5 []
dtype: object
Let’s break down that regex:
Part |
Description |
|---|---|
|
Specifies the start of a string |
|
Square brackets match a single character. When |
|
|
|
|
Regex can do some truly magical things so keep it in mind when you’re doing complicated text wrangling. Let’s see one more example on the cycling dataset:
df = pd.read_csv('data/cycling_data.csv', index_col=0)
df
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 10 Sep 2019, 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 10 Sep 2019, 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 11 Sep 2019, 00:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 11 Sep 2019, 14:06:19 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 12 Sep 2019, 00:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 4 Oct 2019, 01:08:08 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 9 Oct 2019, 13:55:40 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 10 Oct 2019, 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 10 Oct 2019, 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 11 Oct 2019, 00:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
33 rows × 5 columns
We could find all the comments that contains the string “Rain” or “rain”:
df.loc[df['Comments'].str.contains(r"[Rr]ain")]
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 10 Sep 2019, 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 10 Sep 2019, 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 17 Sep 2019, 13:43:34 | Morning Ride | Ride | 2285 | 12.60 | Raining |
| 18 Sep 2019, 13:49:53 | Morning Ride | Ride | 2903 | 14.57 | Raining today |
| 26 Sep 2019, 00:13:33 | Afternoon Ride | Ride | 1860 | 12.52 | raining |
If we didn’t want to include “Raining” or “raining”, we could do:
df.loc[df['Comments'].str.contains(r"^[Rr]ain$")]
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 10 Sep 2019, 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 10 Sep 2019, 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
We can even split strings and separate them into new columns, for example, based on punctuation:
df['Comments'].str.split(r"[.,!]", expand=True)
| 0 | 1 | |
|---|---|---|
| Date | ||
| 10 Sep 2019, 00:13:04 | Rain | None |
| 10 Sep 2019, 13:52:18 | rain | None |
| 11 Sep 2019, 00:23:50 | Wet road but nice weather | None |
| 11 Sep 2019, 14:06:19 | Stopped for photo of sunrise | None |
| 12 Sep 2019, 00:28:05 | Tired by the end of the week | None |
| ... | ... | ... |
| 4 Oct 2019, 01:08:08 | Very tired | riding into the wind |
| 9 Oct 2019, 13:55:40 | Really cold | But feeling good |
| 10 Oct 2019, 00:10:31 | Feeling good after a holiday break | |
| 10 Oct 2019, 13:47:14 | Stopped for photo of sunrise | None |
| 11 Oct 2019, 00:16:57 | Bike feeling tight | needs an oil and pump |
33 rows × 2 columns
My point being here that you can pretty much do anything your heart desires!
2. Working With Datetimes¶
Just like with strings, Pandas has extensive functionality for working with time series data.
Datetime dtype and Motivation for Using Pandas¶
Python has built-in support for datetime format, that is, an object that contains time and date information, in the datetime module.
from datetime import datetime, timedelta
date = datetime(year=2005, month=7, day=9, hour=13, minute=54)
date
datetime.datetime(2005, 7, 9, 13, 54)
We can also parse directly from a string, see format codes here:
date = datetime.strptime("July 9 2005, 13:54", "%B %d %Y, %H:%M")
date
datetime.datetime(2005, 7, 9, 13, 54)
We can then extract specific information from our data:
print(f"Year: {date.strftime('%Y')}")
print(f"Month: {date.strftime('%B')}")
print(f"Day: {date.strftime('%d')}")
print(f"Day name: {date.strftime('%A')}")
print(f"Day of year: {date.strftime('%j')}")
print(f"Time of day: {date.strftime('%p')}")
Year: 2005
Month: July
Day: 09
Day name: Saturday
Day of year: 190
Time of day: PM
And perform basic operations, like adding a week:
date + timedelta(days=7)
datetime.datetime(2005, 7, 16, 13, 54)
But as with strings, working with arrays of datetimes in Python can be difficult and inefficient. NumPy, therefore included a new datetime object to work more effectively with dates:
dates = np.array(["2020-07-09", "2020-08-10"], dtype="datetime64")
dates
array(['2020-07-09', '2020-08-10'], dtype='datetime64[D]')
We can create arrays using other built-in functions like np.arange() too:
dates = np.arange("2020-07", "2020-12", dtype='datetime64[M]')
dates
array(['2020-07', '2020-08', '2020-09', '2020-10', '2020-11'],
dtype='datetime64[M]')
Now we can easily do operations on arrays of time. You can check out all the datetime units and their format in the documentation here.
dates + np.timedelta64(2, 'M')
array(['2020-09', '2020-10', '2020-11', '2020-12', '2021-01'],
dtype='datetime64[M]')
But while numpy helps bring datetimes into the array world, it’s missing a lot of functionality that we would commonly want/need for wrangling tasks. This is where Pandas comes in. Pandas consolidates and extends functionality from the datetime module, numpy, and other libraries like scikits.timeseries into a single place. Pandas provides 4 key datetime objects which we’ll explore in the following sections:
Timestamp (like np.datetime64)
Timedelta (like np.timedelta64)
Period (custom object for regular ranges of datetimes)
DateOffset (custom object like timedelta but factoring in calendar rules)
Creating Datetimes¶
From scratch¶
Most commonly you’ll want to:
Create a single point in time with
pd.Timestamp(), e.g.,2005-07-09 00:00:00Create a span of time with
pd.Period(), e.g.,2020 JanCreate an array of datetimes with
pd.date_range()orpd.period_range()
print(pd.Timestamp('2005-07-09')) # parsed from string
print(pd.Timestamp(year=2005, month=7, day=9)) # pass data directly
print(pd.Timestamp(datetime(year=2005, month=7, day=9))) # from datetime object
2005-07-09 00:00:00
2005-07-09 00:00:00
2005-07-09 00:00:00
The above is a specific point in time. Below, we can use pd.Period() to specify a span of time (like a day):
span = pd.Period('2005-07-09')
print(span)
print(span.start_time)
print(span.end_time)
2005-07-09
2005-07-09 00:00:00
2005-07-09 23:59:59.999999999
point = pd.Timestamp('2005-07-09 12:00')
span = pd.Period('2005-07-09')
print(f"Point: {point}")
print(f" Span: {span}")
print(f"Point in span? {span.start_time < point < span.end_time}")
Point: 2005-07-09 12:00:00
Span: 2005-07-09
Point in span? True
Often, you’ll want to create arrays of datetimes, not just single values. Arrays of datetimes are of the class DatetimeIndex/PeriodIndex/TimedeltaIndex:
pd.date_range('2020-09-01 12:00',
'2020-09-11 12:00',
freq='D')
DatetimeIndex(['2020-09-01 12:00:00', '2020-09-02 12:00:00',
'2020-09-03 12:00:00', '2020-09-04 12:00:00',
'2020-09-05 12:00:00', '2020-09-06 12:00:00',
'2020-09-07 12:00:00', '2020-09-08 12:00:00',
'2020-09-09 12:00:00', '2020-09-10 12:00:00',
'2020-09-11 12:00:00'],
dtype='datetime64[ns]', freq='D')
pd.period_range('2020-09-01',
'2020-09-11',
freq='D')
PeriodIndex(['2020-09-01', '2020-09-02', '2020-09-03', '2020-09-04',
'2020-09-05', '2020-09-06', '2020-09-07', '2020-09-08',
'2020-09-09', '2020-09-10', '2020-09-11'],
dtype='period[D]', freq='D')
We can use Timedelta objects to perform temporal operations like adding or subtracting time:
pd.date_range('2020-09-01 12:00', '2020-09-11 12:00', freq='D') + pd.Timedelta('1.5 hour')
DatetimeIndex(['2020-09-01 13:30:00', '2020-09-02 13:30:00',
'2020-09-03 13:30:00', '2020-09-04 13:30:00',
'2020-09-05 13:30:00', '2020-09-06 13:30:00',
'2020-09-07 13:30:00', '2020-09-08 13:30:00',
'2020-09-09 13:30:00', '2020-09-10 13:30:00',
'2020-09-11 13:30:00'],
dtype='datetime64[ns]', freq='D')
Finally, Pandas represents missing datetimes with NaT, which is just like np.nan:
pd.Timestamp(pd.NaT)
NaT
By converting existing data¶
It’s fairly common to have an array of dates as strings. We can use pd.to_datetime() to convert these to datetime:
string_dates = ['July 9, 2020', 'August 1, 2020', 'August 28, 2020']
string_dates
['July 9, 2020', 'August 1, 2020', 'August 28, 2020']
pd.to_datetime(string_dates)
DatetimeIndex(['2020-07-09', '2020-08-01', '2020-08-28'], dtype='datetime64[ns]', freq=None)
For more complex datetime format, use the format argument (see Python Format Codes for help):
string_dates = ['2020 9 July', '2020 1 August', '2020 28 August']
pd.to_datetime(string_dates, format="%Y %d %B")
DatetimeIndex(['2020-07-09', '2020-08-01', '2020-08-28'], dtype='datetime64[ns]', freq=None)
Or use a dictionary:
dict_dates = pd.to_datetime({"year": [2020, 2020, 2020],
"month": [7, 8, 8],
"day": [9, 1, 28]}) # note this is a series, not an index!
dict_dates
0 2020-07-09
1 2020-08-01
2 2020-08-28
dtype: datetime64[ns]
pd.Index(dict_dates)
DatetimeIndex(['2020-07-09', '2020-08-01', '2020-08-28'], dtype='datetime64[ns]', freq=None)
By reading directly from an external source¶
Let’s practice by reading in our favourite cycling dataset:
df = pd.read_csv('data/cycling_data.csv', index_col=0)
df
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 10 Sep 2019, 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 10 Sep 2019, 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 11 Sep 2019, 00:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 11 Sep 2019, 14:06:19 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 12 Sep 2019, 00:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 4 Oct 2019, 01:08:08 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 9 Oct 2019, 13:55:40 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 10 Oct 2019, 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 10 Oct 2019, 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 11 Oct 2019, 00:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
33 rows × 5 columns
Our index is just a plain old index at the moment, with dtype object, full of string dates:
print(df.index.dtype)
type(df.index)
object
pandas.core.indexes.base.Index
We could manually convert our index to a datetime using pd.to_datetime(). But even better, pd.read_csv() has an argument parse_dates which can do this automatically when reading the file:
df = pd.read_csv('data/cycling_data.csv', index_col=0, parse_dates=True)
df
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-09-10 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 2019-09-10 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 2019-09-11 00:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 2019-09-11 14:06:19 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 2019-09-12 00:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 2019-10-04 01:08:08 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 2019-10-09 13:55:40 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 2019-10-10 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-10 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 2019-10-11 00:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
33 rows × 5 columns
type(df.index)
pandas.core.indexes.datetimes.DatetimeIndex
print(df.index.dtype)
type(df.index)
datetime64[ns]
pandas.core.indexes.datetimes.DatetimeIndex
The parse_dates argument is very flexible and you can specify the datetime format for harder to read dates. There are other related arguments like date_parser, dayfirst, etc that are also helpful, check out the Pandas documentation for more.
Indexing Datetimes¶
Datetime index objects are just like regular Index objects and can be selected, sliced, filtered, etc.
df
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-09-10 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 2019-09-10 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 2019-09-11 00:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 2019-09-11 14:06:19 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 2019-09-12 00:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 2019-10-04 01:08:08 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 2019-10-09 13:55:40 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 2019-10-10 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-10 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 2019-10-11 00:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
33 rows × 5 columns
We can do partial string indexing:
df.loc['2019-09']
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-09-10 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 2019-09-10 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 2019-09-11 00:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 2019-09-11 14:06:19 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 2019-09-12 00:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 2019-09-25 13:35:41 | Morning Ride | Ride | 2124 | 12.65 | Stopped for photo of sunrise |
| 2019-09-26 00:13:33 | Afternoon Ride | Ride | 1860 | 12.52 | raining |
| 2019-09-26 13:42:43 | Morning Ride | Ride | 2350 | 12.91 | Detour around trucks at Jericho |
| 2019-09-27 01:00:18 | Afternoon Ride | Ride | 1712 | 12.47 | Tired by the end of the week |
| 2019-09-30 13:53:52 | Morning Ride | Ride | 2118 | 12.71 | Rested after the weekend! |
22 rows × 5 columns
Exact matching:
df.loc['2019-10-10']
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-10-10 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-10 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
df.loc['2019-10-10 13:47:14']
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-10-10 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
And slicing:
df.loc['2019-10-01':'2019-10-13']
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-10-01 00:15:07 | Afternoon Ride | Ride | 1732 | NaN | Legs feeling strong! |
| 2019-10-01 13:45:55 | Morning Ride | Ride | 2222 | 12.82 | Beautiful morning! Feeling fit |
| 2019-10-02 00:13:09 | Afternoon Ride | Ride | 1756 | NaN | A little tired today but good weather |
| 2019-10-02 13:46:06 | Morning Ride | Ride | 2134 | 13.06 | Bit tired today but good weather |
| 2019-10-03 00:45:22 | Afternoon Ride | Ride | 1724 | 12.52 | Feeling good |
| 2019-10-03 13:47:36 | Morning Ride | Ride | 2182 | 12.68 | Wet road |
| 2019-10-04 01:08:08 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 2019-10-09 13:55:40 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 2019-10-10 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-10 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 2019-10-11 00:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
df.query() will also work here:
df.query("'2019-10-10'")
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-10-10 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-10 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
And for getting all results between two times of a day, use df.between_time():
df.between_time('00:00', '01:00')
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-09-10 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 2019-09-11 00:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 2019-09-12 00:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| 2019-09-17 00:15:47 | Afternoon Ride | Ride | 1973 | 12.45 | Legs feeling strong! |
| 2019-09-18 00:15:52 | Afternoon Ride | Ride | 2101 | 12.48 | Pumped up tires |
| 2019-09-19 00:30:01 | Afternoon Ride | Ride | 48062 | 12.48 | Feeling good |
| 2019-09-24 00:35:42 | Afternoon Ride | Ride | 2076 | 12.47 | Oiled chain, bike feels smooth |
| 2019-09-25 00:07:21 | Afternoon Ride | Ride | 1775 | 12.10 | Feeling really tired |
| 2019-09-26 00:13:33 | Afternoon Ride | Ride | 1860 | 12.52 | raining |
| 2019-10-01 00:15:07 | Afternoon Ride | Ride | 1732 | NaN | Legs feeling strong! |
| 2019-10-02 00:13:09 | Afternoon Ride | Ride | 1756 | NaN | A little tired today but good weather |
| 2019-10-03 00:45:22 | Afternoon Ride | Ride | 1724 | 12.52 | Feeling good |
| 2019-10-10 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-11 00:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
For more complicated filtering, we may have to decompose our timeseries, as we’ll shown in the next section.
Manipulating Datetimes¶
Decomposition¶
We can easily decompose our timeseries into its constituent components. There are many attributes that define these constituents.
df.index.year
Int64Index([2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019,
2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019, 2019],
dtype='int64', name='Date')
df.index.second
Int64Index([ 4, 18, 50, 19, 5, 48, 47, 34, 53, 52, 1, 9, 5, 41, 42, 24, 21,
41, 33, 43, 18, 52, 7, 55, 9, 6, 22, 36, 8, 40, 31, 14, 57],
dtype='int64', name='Date')
df.index.weekday
Int64Index([1, 1, 2, 2, 3, 0, 1, 1, 2, 2, 3, 3, 4, 0, 1, 1, 2, 2, 3, 3, 4, 0,
1, 1, 2, 2, 3, 3, 4, 2, 3, 3, 4],
dtype='int64', name='Date')
As well as methods we can use:
df.index.day_name()
Index(['Tuesday', 'Tuesday', 'Wednesday', 'Wednesday', 'Thursday', 'Monday',
'Tuesday', 'Tuesday', 'Wednesday', 'Wednesday', 'Thursday', 'Thursday',
'Friday', 'Monday', 'Tuesday', 'Tuesday', 'Wednesday', 'Wednesday',
'Thursday', 'Thursday', 'Friday', 'Monday', 'Tuesday', 'Tuesday',
'Wednesday', 'Wednesday', 'Thursday', 'Thursday', 'Friday', 'Wednesday',
'Thursday', 'Thursday', 'Friday'],
dtype='object', name='Date')
df.index.month_name()
Index(['September', 'September', 'September', 'September', 'September',
'September', 'September', 'September', 'September', 'September',
'September', 'September', 'September', 'September', 'September',
'September', 'September', 'September', 'September', 'September',
'September', 'September', 'October', 'October', 'October', 'October',
'October', 'October', 'October', 'October', 'October', 'October',
'October'],
dtype='object', name='Date')
Note that if you’re operating on a Series rather than a DatetimeIndex object, you can access this functionality through the .dt attribute:
s = pd.Series(pd.date_range('2011-12-29', '2011-12-31'))
s.year # raises error
---------------------------------------------------------------------------
AttributeError Traceback (most recent call last)
<ipython-input-60-e24fea3644a8> in <module>
1 s = pd.Series(pd.date_range('2011-12-29', '2011-12-31'))
----> 2 s.year # raises error
/opt/miniconda3/envs/mds511/lib/python3.7/site-packages/pandas/core/generic.py in __getattr__(self, name)
5128 if self._info_axis._can_hold_identifiers_and_holds_name(name):
5129 return self[name]
-> 5130 return object.__getattribute__(self, name)
5131
5132 def __setattr__(self, name: str, value) -> None:
AttributeError: 'Series' object has no attribute 'year'
s.dt.year # works
0 2011
1 2011
2 2011
dtype: int64
Offsets and Timezones¶
We saw before how we can use Timedelta to add/subtract time to our datetimes. Timedelta respects absolute time, which can be problematic in some cases, where time is not regular. For example, on March 8, Canada daylight savings started and clocks moved forward 1 hour. This extra “calendar hour” is not accounted for in absolute time:
t1 = pd.Timestamp('2020-03-07 12:00:00', tz='Canada/Pacific')
t2 = t1 + pd.Timedelta("1 day")
print(f"Original time: {t1}")
print(f" Plus one day: {t2}") # note that time has moved from 12:00 -> 13:00
Original time: 2020-03-07 12:00:00-08:00
Plus one day: 2020-03-08 13:00:00-07:00
Instead, we’d need to use a Dateoffset:
t3 = t1 + pd.DateOffset(days=1)
print(f"Original time: {t1}")
print(f" Plus one day: {t3}") # note that time has stayed at 12:00
Original time: 2020-03-07 12:00:00-08:00
Plus one day: 2020-03-08 12:00:00-07:00
You can see that we started including timezone information above. By default, datetime objects are “timezone unaware”. To associate times with a timezone, we can use the tz argument in construction, or we can use the tz_localize() method:
print(f" No timezone: {pd.Timestamp('2020-03-07 12:00:00').tz}")
print(f" tz arg: {pd.Timestamp('2020-03-07 12:00:00', tz='Canada/Pacific').tz}")
print(f".tz_localize method: {pd.Timestamp('2020-03-07 12:00:00').tz_localize('Canada/Pacific').tz}")
No timezone: None
tz arg: Canada/Pacific
.tz_localize method: Canada/Pacific
You can convert between timezones using the .tz_convert() method. You might have noticed something funny about the times I’ve been riding to University:
df = pd.read_csv('data/cycling_data.csv', index_col=0, parse_dates=True)
df
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-09-10 00:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 2019-09-10 13:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 2019-09-11 00:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 2019-09-11 14:06:19 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 2019-09-12 00:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 2019-10-04 01:08:08 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 2019-10-09 13:55:40 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 2019-10-10 00:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-10 13:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 2019-10-11 00:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
33 rows × 5 columns
I know for a fact that I haven’t been cycling around midnight… There’s something wrong with the timezone in this dataset. I was using the Strava app to document my rides, it was recording in Canadian time but converting to Australia time. Let’s go ahead and fix that up:
df.index = df.index.tz_localize("Canada/Pacific") # first specify the current timezone
df.index = df.index.tz_convert("Australia/Sydney") # then convert to the proper timezone
df
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-09-10 17:13:04+10:00 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 2019-09-11 06:52:18+10:00 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 2019-09-11 17:23:50+10:00 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 2019-09-12 07:06:19+10:00 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 2019-09-12 17:28:05+10:00 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 2019-10-04 18:08:08+10:00 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 2019-10-10 07:55:40+11:00 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 2019-10-10 18:10:31+11:00 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-11 07:47:14+11:00 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 2019-10-11 18:16:57+11:00 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
33 rows × 5 columns
We could have also used a DateOffset if we knew the offset we wanted to apply, in this case, 7 hours:
df = pd.read_csv('data/cycling_data.csv', index_col=0, parse_dates=True)
df.index = df.index + pd.DateOffset(hours=-7)
df
| Name | Type | Time | Distance | Comments | |
|---|---|---|---|---|---|
| Date | |||||
| 2019-09-09 17:13:04 | Afternoon Ride | Ride | 2084 | 12.62 | Rain |
| 2019-09-10 06:52:18 | Morning Ride | Ride | 2531 | 13.03 | rain |
| 2019-09-10 17:23:50 | Afternoon Ride | Ride | 1863 | 12.52 | Wet road but nice weather |
| 2019-09-11 07:06:19 | Morning Ride | Ride | 2192 | 12.84 | Stopped for photo of sunrise |
| 2019-09-11 17:28:05 | Afternoon Ride | Ride | 1891 | 12.48 | Tired by the end of the week |
| ... | ... | ... | ... | ... | ... |
| 2019-10-03 18:08:08 | Afternoon Ride | Ride | 1870 | 12.63 | Very tired, riding into the wind |
| 2019-10-09 06:55:40 | Morning Ride | Ride | 2149 | 12.70 | Really cold! But feeling good |
| 2019-10-09 17:10:31 | Afternoon Ride | Ride | 1841 | 12.59 | Feeling good after a holiday break! |
| 2019-10-10 06:47:14 | Morning Ride | Ride | 2463 | 12.79 | Stopped for photo of sunrise |
| 2019-10-10 17:16:57 | Afternoon Ride | Ride | 1843 | 11.79 | Bike feeling tight, needs an oil and pump |
33 rows × 5 columns
Resampling and Aggregating¶
One of the most common operations you will want do when working with time series is resampling the time series to a coarser/finer/regular resolution. For example, you may want to resample daily data to weekly data. We can do that with the .resample() method. For example, let’s resample my irregular cycling timeseries to a regular 12-hourly series:
df.index
DatetimeIndex(['2019-09-09 17:13:04', '2019-09-10 06:52:18',
'2019-09-10 17:23:50', '2019-09-11 07:06:19',
'2019-09-11 17:28:05', '2019-09-16 06:57:48',
'2019-09-16 17:15:47', '2019-09-17 06:43:34',
'2019-09-18 06:49:53', '2019-09-17 17:15:52',
'2019-09-18 17:30:01', '2019-09-19 06:52:09',
'2019-09-19 18:02:05', '2019-09-23 06:50:41',
'2019-09-23 17:35:42', '2019-09-24 06:41:24',
'2019-09-24 17:07:21', '2019-09-25 06:35:41',
'2019-09-25 17:13:33', '2019-09-26 06:42:43',
'2019-09-26 18:00:18', '2019-09-30 06:53:52',
'2019-09-30 17:15:07', '2019-10-01 06:45:55',
'2019-10-01 17:13:09', '2019-10-02 06:46:06',
'2019-10-02 17:45:22', '2019-10-03 06:47:36',
'2019-10-03 18:08:08', '2019-10-09 06:55:40',
'2019-10-09 17:10:31', '2019-10-10 06:47:14',
'2019-10-10 17:16:57'],
dtype='datetime64[ns]', name='Date', freq=None)
df.resample("1D")
<pandas.core.resample.DatetimeIndexResampler object at 0x117e1fb50>
Resampler objects are very similar to the groupby objects we saw in the previous chapter. We need to apply an aggregating function on our grouped timeseries, just like we did with groupby objects:
dfr = df.resample("1D").mean()
dfr
| Time | Distance | |
|---|---|---|
| Date | ||
| 2019-09-09 | 2084.0 | 12.620 |
| 2019-09-10 | 2197.0 | 12.775 |
| 2019-09-11 | 2041.5 | 12.660 |
| 2019-09-12 | NaN | NaN |
| 2019-09-13 | NaN | NaN |
| ... | ... | ... |
| 2019-10-06 | NaN | NaN |
| 2019-10-07 | NaN | NaN |
| 2019-10-08 | NaN | NaN |
| 2019-10-09 | 1995.0 | 12.645 |
| 2019-10-10 | 2153.0 | 12.290 |
32 rows × 2 columns
There’s quite a few NaNs in there? Some days I didn’t ride, but some might by weekends too…
dfr['Weekday'] = dfr.index.day_name()
dfr.head(10)
| Time | Distance | Weekday | |
|---|---|---|---|
| Date | |||
| 2019-09-09 | 2084.0 | 12.620 | Monday |
| 2019-09-10 | 2197.0 | 12.775 | Tuesday |
| 2019-09-11 | 2041.5 | 12.660 | Wednesday |
| 2019-09-12 | NaN | NaN | Thursday |
| 2019-09-13 | NaN | NaN | Friday |
| 2019-09-14 | NaN | NaN | Saturday |
| 2019-09-15 | NaN | NaN | Sunday |
| 2019-09-16 | 2122.5 | 12.450 | Monday |
| 2019-09-17 | 2193.0 | 12.540 | Tuesday |
| 2019-09-18 | 25482.5 | 13.525 | Wednesday |
Pandas support “business time” operations and format codes in all the timeseries functions we’ve seen so far. You can check out the documentation for more info, but let’s specify business days here to get rid of those weekends:
dfr = df.resample("1B").mean() # "B" is business day
dfr['Weekday'] = dfr.index.day_name()
dfr.head(10)
| Time | Distance | Weekday | |
|---|---|---|---|
| Date | |||
| 2019-09-09 | 2084.0 | 12.620 | Monday |
| 2019-09-10 | 2197.0 | 12.775 | Tuesday |
| 2019-09-11 | 2041.5 | 12.660 | Wednesday |
| 2019-09-12 | NaN | NaN | Thursday |
| 2019-09-13 | NaN | NaN | Friday |
| 2019-09-16 | 2122.5 | 12.450 | Monday |
| 2019-09-17 | 2193.0 | 12.540 | Tuesday |
| 2019-09-18 | 25482.5 | 13.525 | Wednesday |
| 2019-09-19 | 2525.5 | 12.700 | Thursday |
| 2019-09-20 | NaN | NaN | Friday |
3. Hierachical Indexing¶
Hierachical indexing, sometimes called “multi-indexing” or “stacked indexing”, is how Pandas “nests” data. The idea is to facilitate the storage of high dimensional data in a 2D dataframe.

Source: Giphy
Creating a Hierachical Index¶
Let’s start with a motivating example. Say you want to track how many courses each Master of Data Science instructor taught over the years in a Pandas Series.
Note
Recall that the content of this site is adapted from material I used to teach the 2020/2021 offering of the course “DSCI 511 Python Programming for Data Science” for the University of British Columbia’s Master of Data Science Program.
We could use a tuple to make an appropriate index:
index = [('Tom', 2019), ('Tom', 2020),
('Mike', 2019), ('Mike', 2020),
('Tiffany', 2019), ('Tiffany', 2020)]
courses = [4, 6, 5, 5, 6, 3]
s = pd.Series(courses, index)
s
(Tom, 2019) 4
(Tom, 2020) 6
(Mike, 2019) 5
(Mike, 2020) 5
(Tiffany, 2019) 6
(Tiffany, 2020) 3
dtype: int64
We can still kind of index this series:
s.loc[("Tom", 2019):("Tom", 2019)]
(Tom, 2019) 4
dtype: int64
But if we wanted to get all of the values for 2019, we’d need to do some messy looping:
s[[i for i in s.index if i[1] == 2019]]
(Tom, 2019) 4
(Mike, 2019) 5
(Tiffany, 2019) 6
dtype: int64
The better way to set up this problem is with a multi-index (“hierachical index”). We can create a multi-index with pd.MultiIndex.from_tuple(). There are other variations of .from_X but tuple is most common.
mi = pd.MultiIndex.from_tuples(index)
mi
MultiIndex([( 'Tom', 2019),
( 'Tom', 2020),
( 'Mike', 2019),
( 'Mike', 2020),
('Tiffany', 2019),
('Tiffany', 2020)],
)
s = pd.Series(courses, mi)
s
Tom 2019 4
2020 6
Mike 2019 5
2020 5
Tiffany 2019 6
2020 3
dtype: int64
Now we can do more efficient and logical indexing:
s.loc['Tom']
2019 4
2020 6
dtype: int64
s.loc[:, 2019]
Tom 4
Mike 5
Tiffany 6
dtype: int64
s.loc["Tom", 2019]
4
We could also create the index by passing iterables like a list of lists directly to the index argument, but I feel it’s not as explicit or intutitive as using pd.MultIndex:
index = [['Tom', 'Tom', 'Mike', 'Mike', 'Tiffany', 'Tiffany'],
[2019, 2020, 2019, 2020, 2019, 2020]]
courses = [4, 6, 5, 5, 6, 3]
s = pd.Series(courses, index)
s
Tom 2019 4
2020 6
Mike 2019 5
2020 5
Tiffany 2019 6
2020 3
dtype: int64
Stacking / Unstacking¶
You might have noticed that we could also represent our multi-index series as a dataframe. Pandas noticed this too and provides the .stack() and .unstack() methods for switching between dataframes and multi-index series:
s = s.unstack()
s
| 2019 | 2020 | |
|---|---|---|
| Mike | 5 | 5 |
| Tiffany | 6 | 3 |
| Tom | 4 | 6 |
s.stack()
Mike 2019 5
2020 5
Tiffany 2019 6
2020 3
Tom 2019 4
2020 6
dtype: int64
Using a Hierachical Index¶
Observing the multi-index <-> dataframe equivalence above, you might wonder why we would even want multi-indices. Above, we were only dealing with 2D data, but a multi-index allows us to store any arbitrary number of dimensions:
index = [['Tom', 'Tom', 'Mike', 'Mike', 'Tiffany', 'Tiffany'],
[2019, 2020, 2019, 2020, 2019, 2020]]
courses = [4, 6, 5, 5, 6, 3]
s = pd.Series(courses, index)
s
Tom 2019 4
2020 6
Mike 2019 5
2020 5
Tiffany 2019 6
2020 3
dtype: int64
pd.DataFrame(s).stack()
Tom 2019 0 4
2020 0 6
Mike 2019 0 5
2020 0 5
Tiffany 2019 0 6
2020 0 3
dtype: int64
s.loc['Tom']
2019 4
2020 6
dtype: int64
tom = pd.DataFrame({"Courses": [4, 6],
"Students": [273, 342]},
index = [2019, 2020])
mike = pd.DataFrame({"Courses": [5, 5],
"Students": [293, 420]},
index = [2019, 2020])
tiff = pd.DataFrame({"Courses": [6, 3],
"Students": [363, 190]},
index = [2019, 2020])
Here I have three 2D dataframes that I’d like to join together. There are so many ways you can do this, but I’m going to use pd.concat() and then specify the keys argument:
s3 = pd.concat((tom, mike, tiff),
keys= ['Tom', 'Mike', 'Tiff'],
axis=0)
s3
| Courses | Students | ||
|---|---|---|---|
| Tom | 2019 | 4 | 273 |
| 2020 | 6 | 342 | |
| Mike | 2019 | 5 | 293 |
| 2020 | 5 | 420 | |
| Tiff | 2019 | 6 | 363 |
| 2020 | 3 | 190 |
Now we have 3 dimensions of information in a single structure!
s3.stack()
Tom 2019 Courses 4
Students 273
2020 Courses 6
Students 342
Mike 2019 Courses 5
Students 293
2020 Courses 5
Students 420
Tiff 2019 Courses 6
Students 363
2020 Courses 3
Students 190
dtype: int64
s3.loc['Tom']
| Courses | Students | |
|---|---|---|
| 2019 | 4 | 273 |
| 2020 | 6 | 342 |
s3.loc['Tom', 2019]
Courses 4
Students 273
Name: (Tom, 2019), dtype: int64
We can access deeper levels in various ways:
s3.loc['Tom', 2019]['Courses']
4
s3.loc[('Tom', 2019), 'Courses']
4
s3.loc[('Tom', 2019), 'Courses']
4
If we name our index columns, we can also use .query():
s3 = s3.rename_axis(index=["Name", "Year"])
s3
| Courses | Students | ||
|---|---|---|---|
| Name | Year | ||
| Tom | 2019 | 4 | 273 |
| 2020 | 6 | 342 | |
| Mike | 2019 | 5 | 293 |
| 2020 | 5 | 420 | |
| Tiff | 2019 | 6 | 363 |
| 2020 | 3 | 190 |
s3.query("Year == 2019")
| Courses | Students | ||
|---|---|---|---|
| Name | Year | ||
| Tom | 2019 | 4 | 273 |
| Mike | 2019 | 5 | 293 |
| Tiff | 2019 | 6 | 363 |
Or you might prefer the “stacked” version of our hierachical index:
s3.stack()
Name Year
Tom 2019 Courses 4
Students 273
2020 Courses 6
Students 342
Mike 2019 Courses 5
Students 293
2020 Courses 5
Students 420
Tiff 2019 Courses 6
Students 363
2020 Courses 3
Students 190
dtype: int64
s3.stack().loc[('Tom', 2019, 'Courses')]
4
By the way, we can also use all the previous methods we’ve learned about on hierachical dataframes:
s3.sort_index(ascending=False)
| Courses | Students | ||
|---|---|---|---|
| Name | Year | ||
| Tom | 2020 | 6 | 342 |
| 2019 | 4 | 273 | |
| Tiff | 2020 | 3 | 190 |
| 2019 | 6 | 363 | |
| Mike | 2020 | 5 | 420 |
| 2019 | 5 | 293 |
s3.sort_values(by='Students')
| Courses | Students | ||
|---|---|---|---|
| Name | Year | ||
| Tiff | 2020 | 3 | 190 |
| Tom | 2019 | 4 | 273 |
| Mike | 2019 | 5 | 293 |
| Tom | 2020 | 6 | 342 |
| Tiff | 2019 | 6 | 363 |
| Mike | 2020 | 5 | 420 |
There’s one important exception! We can now specify a level argument to chose which level of our multi-index to apply the function to:
s3.mean()
Courses 4.833333
Students 313.500000
dtype: float64
s3.mean(level='Year')
| Courses | Students | |
|---|---|---|
| Year | ||
| 2019 | 5.000000 | 309.666667 |
| 2020 | 4.666667 | 317.333333 |
4. Visualizing DataFrames¶
Pandas provides a .plot() method on Series and DataFrames which I wanted to show briefly here.
Simple Plots¶
df = pd.read_csv('data/cycling_data.csv', index_col=0, parse_dates=True).dropna()
Let’s go ahead and make a plot of the distances I’ve ridden:
df['Distance'].plot.line();
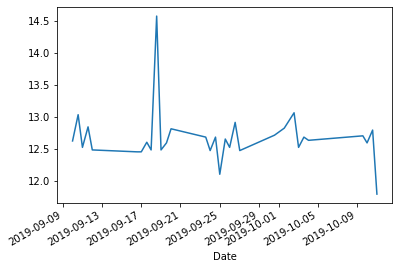
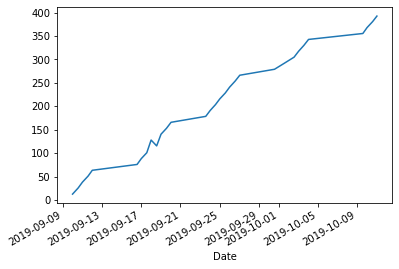
Cumulative distance might be more informative:
df['Distance'].cumsum().plot.line();
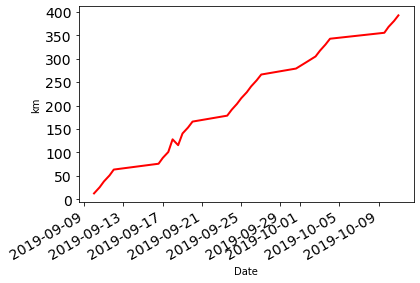
There are many configuration options for these plots which build of the matplotlib library:
df['Distance'].cumsum().plot.line(fontsize=14, linewidth = 2, color = 'r', ylabel="km");
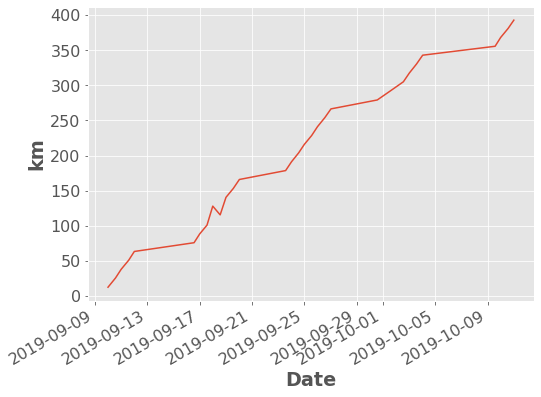
I actually usually use built-in themes for my plots which do a lot of the colour and text formatting for you:
import matplotlib.pyplot as plt
plt.style.use('ggplot')
plt.rcParams.update({'font.size': 16,
'axes.labelweight': 'bold',
'figure.figsize': (8,6)})
df['Distance'].dropna().cumsum().plot.line(ylabel="km");
Some people have also made custom themes, like this fun cyberpunk theme:
import mplcyberpunk
plt.style.use("cyberpunk")
df['Distance'].plot.line(ylabel="km")
mplcyberpunk.add_glow_effects()
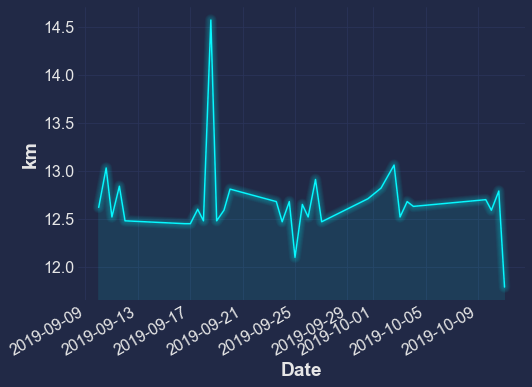
There are many other kinds of plots you can make too:
Method |
Plot Type |
|---|---|
|
bar plots |
|
histogram |
|
boxplot |
|
density plots |
|
area plots |
|
scatter plots |
|
hexagonal bin plots |
|
pie plots |
plt.style.use('ggplot')
plt.rcParams.update({'font.size': 16,
'axes.labelweight': 'bold',
'figure.figsize': (8,6)})
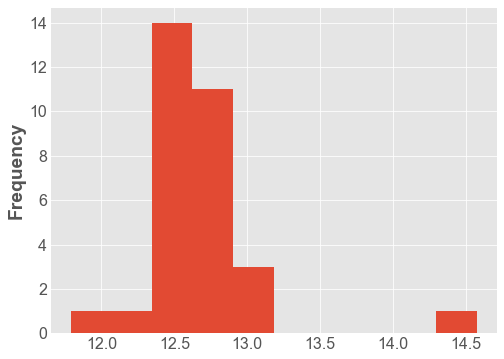
df['Distance'].plot.hist();
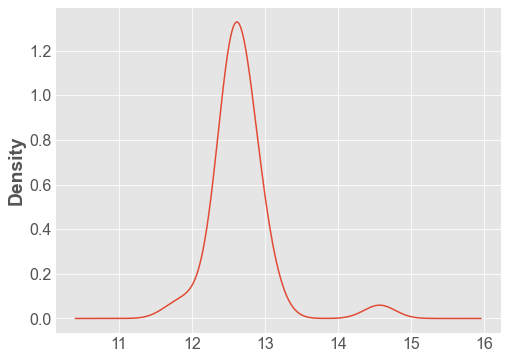
df['Distance'].plot.density();
Pandas Plotting¶
Pandas also supports a few more advanced plotting functions in the pandas.plotting module. You can view them in the Pandas documentation.
from pandas.plotting import scatter_matrix
scatter_matrix(df);
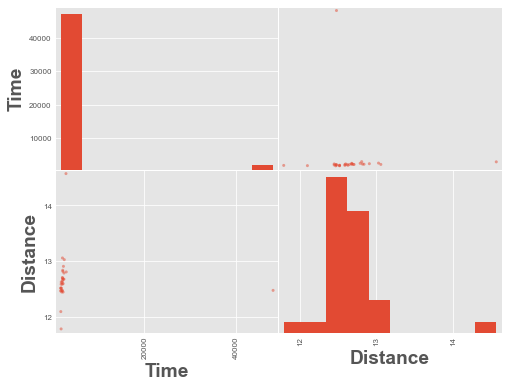
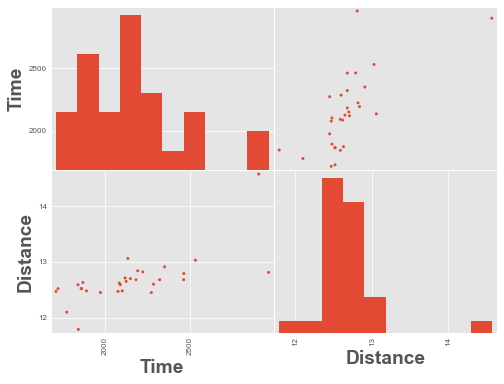
We have an outlier time in the data above, a time value of ~48,000. Let’s remove it and re-plot.
scatter_matrix(df.query('Time < 4000'), alpha=1);
5. Pandas Profiling¶
Pandas profiling is a nifty tool for generating summary reports and doing exploratory data analysis on dataframes. Pandas profiling is not part of base Pandas but you can install with:
$ conda install -c conda-forge pandas-profiling
import pandas_profiling
df = pd.read_csv('data/cycling_data.csv')
df.profile_report(progress_bar=False)
